Projects
flowEMMi
Flow cytometry (FCM) is widely used in health research, diagnostics, biotechnology and environmental microbiology. The technology measures optical properties of millions of cells, which makes FCM uniquely suited to provide a range of basic cell characteristics such as cell sizes, cell densities, and DNA content on both eukaryotic and prokaryotic scales, and in short time frames.
In these figures, taken from Ludwig at al, 2019, we observe localized cell clusters (yellow/red) that we want to mark in a ``gating procedure’’ (left). flowEMMi automatically determines the correct number of gates and performs the gating as can be seen to the right.
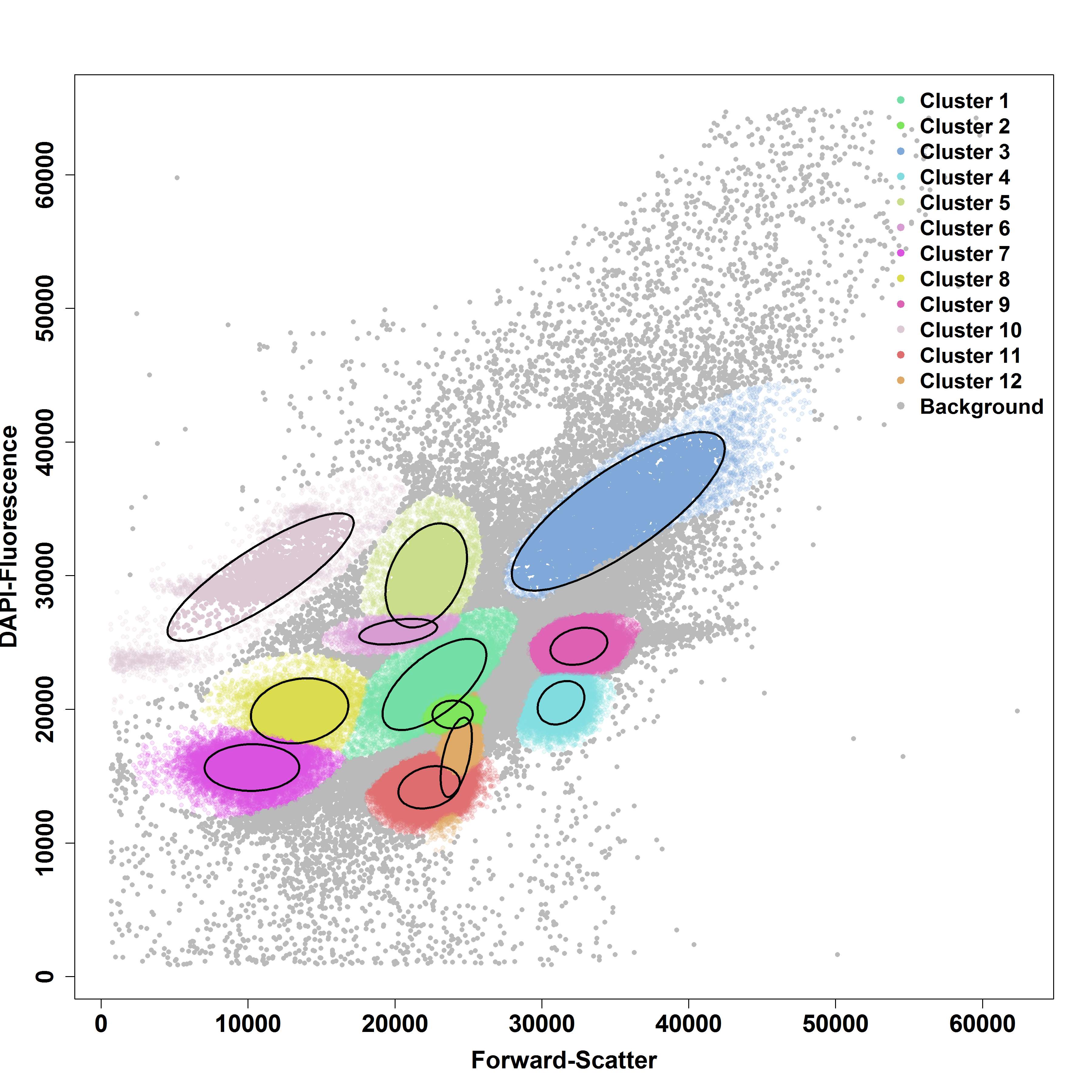
Why is this important?
For cell samples with a widely used background, i.e. human, cell differentiation can be done using labeled antibodies, and different fluorescent excitations. This makes differentiation easy due to labeling of sub-populations with different antibody-coupled fluorescent dyes. Microbial cell types can also be differentiated by antibodies or labeling via FISH (fluorescent in-situ hybridization) but only if the species is available as pure culture or binding sequences are known.
For microbial cells of unknown origin (i.e. from natural microbial communities), this is infeasible as antibodies do not even exist and genome data are not available. Hence, for environmental studies only basic measurements can be provided and cells have to be classified (gated) according to these data only.
ADPfusion
ADPfusion and its related developments provides a framework for dynamic programming (DP) that combines a high level of expressiveness with competitive performance of the resulting dynamic programming algorithms. ADPfusion make liberal use of program fusion to compile programs written in its shallow domain-specific language into efficient program code. ADPfusion is based on well-founded theoretical developments that help the user to compose their DP programs in an algorithmically sound way from more simple building blocks.
We can tackle problems from diverse fields such as sequence alignment, RNA folding, hidden Markov models (HMMs), and scoring of phylogenetic trees. We provide optimal, and suboptimal solutions, stochastic sampling, backtracking, and marginalization or ensemble properties (eg. a posteriori probabilities).
ADPfusion separates state space traversal via linear, context-free, and multiple-context-free grammars, scoring (encoded as an algebra), and choice rule. Our grammars can parse a wide variety of input structures, including strings, trees, sets, and stochastic models.
In this example, taken from Algebraic dynamic programming on trees, Berkemer et al, 2017 we see an (affine) tree alignment program in action, aligning a german to an english sentence.
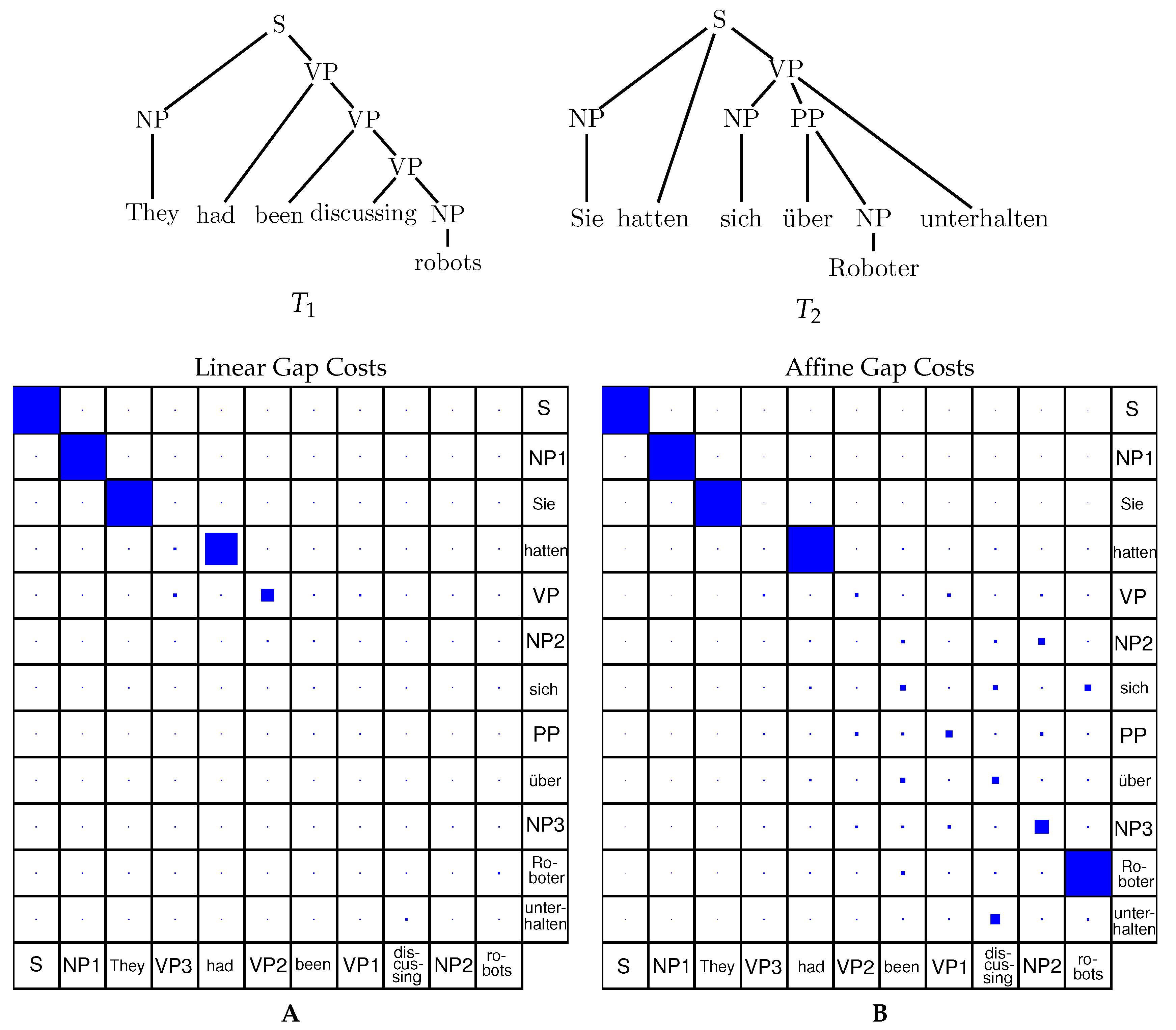
Inferring the (structural) past
Inferring the structural evolution of a non-coding RNA is fraught with complications due to the almost complete absense of “archeologically conserved” structure, with the notable exception of finds like Denisovan where genomic sequences could be extracted. In most cases, one has to begin with only a conserved structure in related species and the extant structure of interest.
We are interested in exact and approximate algorithms that try to recover most likely evolutionary pathways and their structural changes. In Walter Costa et al, 2018 we developed an algorithm that calculates all possible pathways of structural change up to one backmutation and provides evidence for most likely initial, and terminal mutations, as well as pairs coupled of mutations, and relative likelihood of back mutations on certain positions.
This allows to fully explore these pathways and compare them with the (sparse) evidence retrieved from archeological data such as in Denisovan. We can use this algorithm for non-coding sequences that are under repid evolutionary changes compared to other genes.